Get started
The following example briefly demonstrates how PIntron can be used to predict the full-length isoforms of a gene (TP53) starting from its associated UniGene cluster (Hs.437460).
This page is only a brief introduction to PIntron. Please refer to the full documentation for a complete description of its options.
Preparation
Download and install PIntron as described here.
In the rest of this example, we will assume that PIntron executables
have been installed in directory /home/pintron/bin.
Please substitute /home/pintron/bin with the directory where
PIntron has been installed to (or omit the path if PIntron
executables are in a directory listed in $PATH).
Input files
PIntron requires two input files: the
genomic sequence (in this case genomic.txt), and the EST/mRNA
sequences (in this case ests.txt), specified respectively with the
options -g and -s.
File genomic.txt is a FASTA file containing a single sequence, while
ests.txt is a MultiFASTA file, where each sequence is considered as a
single transcript (EST or mRNA).
We strictly require a specific header format.
Please refer to the
documentation for a detailed
description.
The input files for this example are located in the subdirectory
dist-docs/example and are also available for download
(genomic.txt,
ests.txt).
After the preparation, the directory tree should be as follows (other files may exist).
/home/pintron
├── bin
│ ├── cds-annotation
│ ├── compact-compositions
│ ├── est-fact
│ ├── gene-structure
│ ├── intron-agreement
│ ├── maximal-transcripts
│ ├── min-factorization
│ └── pintron
└── doc
└── example
├── ests.txt
└── genomic.txt
Execution
Assuming that we want to generate all output files in the current working directory, the following command executes PIntron on the example.
/home/pintron/bin/pintron \
--bin-dir=/home/pintron/bin \
--genomic=/home/pintron/doc/example/genomic.txt \
--EST=/home/pintron/doc/example/ests.txt \
--organism=human \
--gene=TP53 \
--output=pintron-full-output.json \
--gtf=pintron-cds-annotated-isoforms.gtf \
--extended-gtf=pintron-all-isoforms.gtf \
--logfile=pintron-pipeline-log.txt \
--general-logfile=pintron-log.txt
Most of the options have sensible default values and a short version. Therefore, a shorter equivalent command line is:
/home/pintron/bin/pintron \
-b /home/pintron/bin \
-g /home/pintron/doc/example/genomic.txt \
-s /home/pintron/doc/example/ests.txt \
-n human \
-e TP53
Please notice that options --organism/-n and --gene/-e are optional
and can be omitted (in that case, the default value unknown is assumed).
Output files
PIntron produces the following output files:
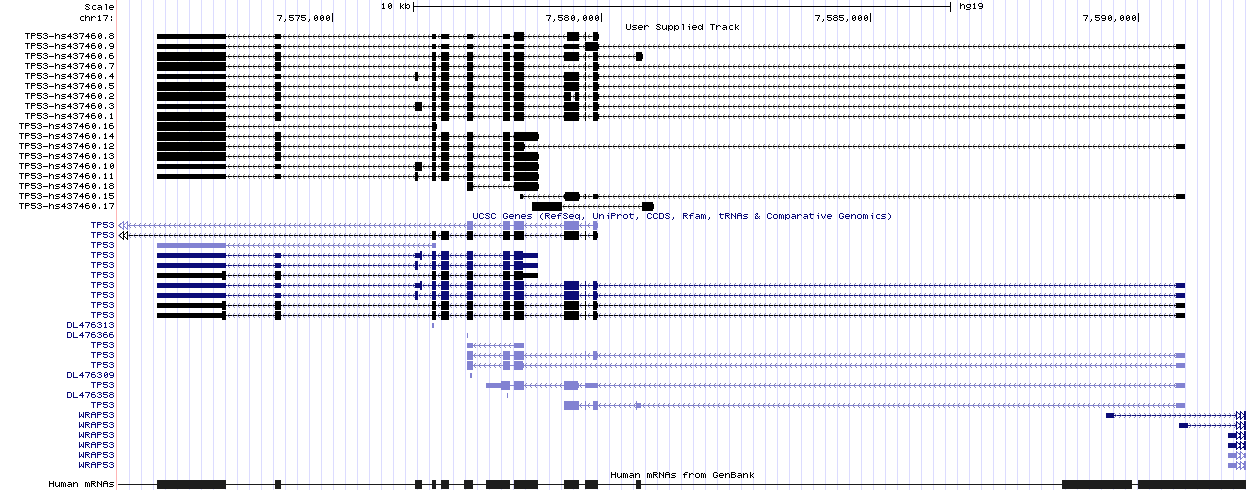
pintron-full-output.json, the complete description of the results computed by PIntron in JSON format. This file is both human- and machine-readable (parsing libraries exist in all the major programming languages). Moreover, the format is almost self-documenting and can be easily adapted to the future needs. Please refer to its description for additional information.pintron-all-isoforms.gtf/pintron-cds-annotated-isoforms.gtf, the set of all (CDS-annotated, respectively) full-length isoforms computed by PIntron in standard GTF2.2 format. These files can be used for some standard downstream analyzes. For example, they can be uploaded to the UCSC Genome Browser as custom tracks (as shown in the example below).pintron-log.txt/pintron-pipeline-log.txt, the logs of main program and of each step of the pipeline. These files could contain important information if an error has occurred. Please upload them with any issue report.
The output files of PIntron on the example gene TP53 are located
in the subdirectory dist-docs/example/sample-output while a graphical
representation of the reconstructed isoforms is as follows: